What is Epigenetics technical articles are geared towards epigenetic research techniques, news, and trends in the field of epigenetics, written by scientists from universities and institutions including UCLA, Hofstra, NIH, Johns Hopkins, and more.
Explore in detail new epigenetic research techniques and tips for topics like next-generation sequencing (NGS) in epigenetics, m6A RNA methylation, CRISPR/Cas9 system epigenetic editing, chromatin immunoprecipitation (ChIP) protocol optimization, and single-cell epigenomics methods.
Want to share your research? Submit your own epigenetics article to be featured.
The concept of DNA library preparation after bisulfite conversion of DNA was originally introduced by Miura et al. (Nucleic Acids Res. 2012) and commercial products for post-bisulfite library preparation were first developed by Epigentek (EpiNext kit) and Epicentre (EpiGnome kit) in 2013, in order to increase sensitivity and reduce sample loss. The major application of post-bisulfite library preparation is whole genome bisulfite sequencing (WGBS) but, with just a slight change to the protocol, this method could also be used for reduced representation [more…]
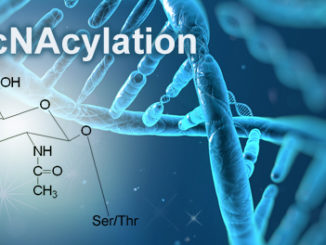
Post-translational modifications are well known for their influence on protein stability, enzymatic functions, as well as protein:protein interactions. At the level of gene expression, acetylation, methylation, phosphorylation, ADP-ribosylation, ubiquitination and SUMOylation are some of the most prevalent chemical modifications that regulate transcription factors and the chromatin template (the complex of DNA and histone proteins) alike. In particular, the manifold modifications occurring on the histone proteins that closely associate with DNA are proposed to constitute a gene-regulating code (the histone code). [more…]
Recent developments in DNA methylation analysis technologies have made it crucial for researchers to understand which tool is optimal for their epigenetic research. These new methods pose exciting opportunities never before imagined, allowing for epigenetic variation to be connected to phenotypic consequences on a much grander scale and at single-base resolution. In a recent issue of an epigenetics newsletter, The Decoder, scientists at Epigentek discuss the latest progress made in profiling genome-wide and region-specific DNA methylation and offer suggestions on [more…]
DNA bisulfite conversion is a unique tool used to discriminate between methylated and unmethylated cytosines for DNA methylation studies. Only bisulfite modification of DNA followed by sequencing yields reliable information on the methylation states of individual cytosines at single base resolution. The bisulfite modification technique uses bisulfite salt to deaminate cytosine residues on single-stranded DNA, converting them to uracil while leaving 5-methylcytosine unchanged (Figure 1). To effectively and efficiently prepare converted DNA for use in various downstream analyses, an ideal [more…]
A new single-cell bisulfite sequencing (scBS-seq) technique that can advance epigenetic experiments has been developed by researchers from BSRC-funded Babraham Institute and the Wellcome Trust Sanger Institute Single Cell Genomics Centre. Using this powerful technique, all epigenetic marks on the DNA within a single cell can be mapped out. This novel method could enhance our understanding of embryonic development and holds promise for improving clinical applications such as fertility treatments and cancer therapy. It may also reduce the amount of [more…]
Over the past decade significant advances have been made in methylation profiling technology allowing for highly specific and accurate information about the epigenome of various species. Because the 5mC and 5hmC modifications are widespread with possibly different functions, further insight into their distribution is important. Traditional methylation analysis methods such as mass spec, HPLC and TLC allow high accuracy but also require sophisticated equipment, are not high through-put and most importantly are expensive. Likewise more modern applications such as next [more…]
Every epigenetics scientist knows that chromatin immunoprecipitation (ChIP) is a valuable technique for studying protein-DNA interactions. They also know that antibodies used in ChIP to capture the DNA/protein complex must be reliable and specifically recognize the fixed protein that is bound to the chromatin complex. But how can a researcher be certain that the antibodies they are using work well in ChIP? For many researchers it is not always the case to be “blessed” with a quality ChIP-grade antibody and, unfortunately, [more…]
Researchers rely on high quality antibodies to perform successful chromatin immunoprecipitation (ChIP) experiments. When investigating the protein-DNA interaction of interest, valid and reliable results simply cannot be attained using nonspecific and inefficient antibodies. If the antibody doesn’t come “ChIP-qualified” or “ChIP-grade” from the supplier, there are several tips you can follow to determine if it’s likely to perform well in ChIP and won’t pull down distracting material to get in the way of a successful study. Carry out the standard [more…]
The following histone extraction protocol was written by scientists at Epigentek and is a recommended and optimized procedure for their histone modification assays, successfully utilized by many research labs. Use these easy to follow steps to ensure proper isolation of histone proteins. For tissues (treated and untreated), weigh the sample and cut the sample into small pieces (1-2 mm3) with a scalpel or scissors. Transfer tissue pieces to a Dounce homogenizer. Add TEB buffer (PBS containing 0.5% Triton X 100, 2 [more…]
Hematopoietic stem cells (HSCs) are rare bone marrow cells that have self-renewal capability and are multipotent. Upon differentiation, HSCs become progressively lineage-committed and give rise to different mature blood cells. This process involves extrinsic and intrinsic signals that are strongly influenced by the stem cell microenvironment. Furthermore, differentiation involves silencing of self-renewal genes and induction of a specific transcriptional program. It is not known how epigenetic modifications influences stem cell differentiation and commitment and what specific role these modifications may [more…]