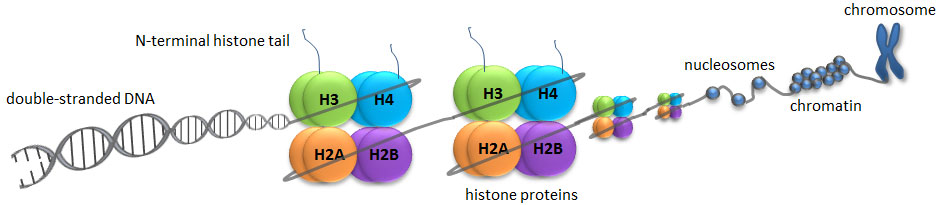
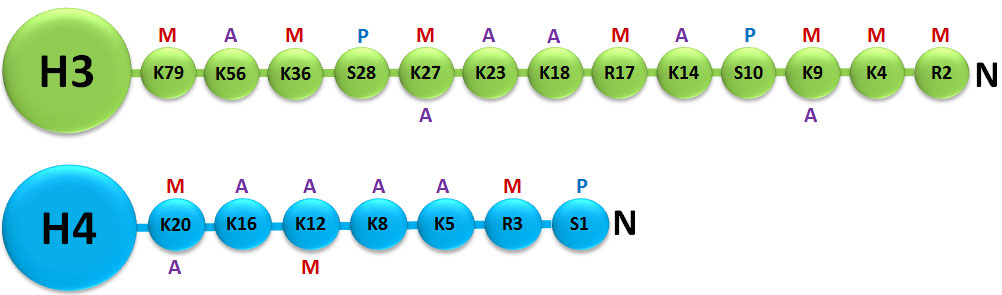
A histone modification is a covalent post-translational modification (PTM) to histone proteins which includes methylation, phosphorylation, acetylation, ubiquitylation, and sumoylation. The PTMs made to histones can impact gene expression by altering chromatin structure or recruiting histone modifiers. Histone proteins act to package DNA, which wraps around the eight histones, into chromosomes. Histone modifications act in diverse biological processes such as transcriptional activation/inactivation, chromosome packaging, and DNA damage/repair. In most species, histone H3 is primarily acetylated at lysines 9, 14, 18, 23, and 56, methylated at arginine 2 and lysines 4, 9, 27, 36, and 79, and phosphorylated at ser10, ser28, Thr3, and Thr11. Histone H4 is primarily acetylated at lysines 5, 8, 12 and 16, methylated at arginine 3 and lysine 20, and phosphorylated at serine 1. Thus, quantitative detection of various histone modifications would provide useful information for a better understanding of epigenetic regulation of cellular processes and the development of histone modifying enzyme-targeted drugs.
Histone Acetylation/Deacetylation
Histone acetylation occurs by the enzymatic addition of an acetyl group (COCH3) from acetyl coenzyme A. The process of histone acetylation is tightly involved in the regulation of many cellular processes including chromatin dynamics and transcription, gene silencing, cell cycle progression, apoptosis, differentiation, DNA replication, DNA repair, nuclear import, and neuronal repression. The modifying enzymes involved in histone acetylation are called histone acetyltransferases (HATs) and they play a critical role in controlling histone H3 and H4 acetylation. More than 20 HATs have been identified which can be classified into five families: GNAT1, MYST, TAFII250, P300/CBP, and nuclear receptor coactivators such as ACTR.1 Histone H3 acetylation may be increased by inhibition of histone deacetylases (HDACs) and decreased by HAT inhibition.
Histone deacetylaces (HDACs) catalyze the hydrolytic removal of acetyl groups from histone lysine residues. An imbalance in the equilibrium of histone acetylation has been associated with tumorigenesis and cancer progression. Detecting whether histone H3 is acetylated at its lysine residues would provide useful information for further characterization of acetylation patterns or sites, thereby leading to a better understanding of epigenetic regulation of gene activation as well as the development of HAT-targeted drugs. Similar to HATs, HDACs play a critical role in various cellular processes involving histone H3 and H4. So far, at least 4 classes of HDACs have been identified. Class I HDACs include 1, 2, 3, and 8. Class II HDACs are comprised of 4, 5, 6, 7, 9, and 10. Class III enzymes, known as sirtuins, require NAD+ cofactors and include SIRTs 1-7. The Class IV enzyme, which contains only HDAC11, has features of both Class I and II. HDAC inhibition displays significant effects on apoptosis, cell cycle arrest, and differentiation in cancer cells. HDAC inhibitors are currently being developed as anticancer agents.2
Extract your nuclear proteins from your sample of interest, then utilize an ELISA-like method to measure the amount of HDAC proteins, or the activity levels of HDAC or activity levels of HAT.
Histone Methylation/Demethylation
Histone methylation is defined as the transfer of one, two, or three methyl groups from S-adenosyl-L-methionine to lysine or arginine residues of histone proteins by histone methyltransferases (HMTs). HMTs control or regulate DNA methylation through chromatin-dependent transcriptional repression or activation. In the cell nucleus, when histone methylation occurs, specific genes within the DNA complexed with the histone may be activated or silenced.3 Several different histone methyltransferases exist that are specific for the lysine or arginine residue which they modify. On histone H3 for example, SET1, SET7/9, Ash1, ALL-1, MLL, ALR, Trx, and SMYD3 are histone methyltransferases that catalyze methylation of histone H3 at lysine 4 (H3-K4) in mammalian cells. ESET, G9a, SUV39-h1, SUV39-h2, SETDB1, Dim-5, and Eu-HMTase are histone methyltransferases that catalyze methylation of histone H3 at lysine 9 (H3-K9) in mammalian cells. G9a and polycomb group enzymes such as EZH2 are histone methyltransferases that catalyze methylation of histone H3 at lysine 27 (H3-K27) in mammalian cells.4 Both H3-K9 and H3-K27 methylation mediates heterochromatin formation and also participates in silencing gene expression at euchromatic sites. Increased global H3-K27 methylation is also found to be involved in some pathological processes such as cancer progression.

