Chromatin remodeling is the rearrangement of chromatin from a condensed state to a transcriptionally accessible state, allowing transcription factors or other DNA binding proteins to access DNA and control gene expression. Chromatin is the complex of DNA and proteins that are packed within the nucleus of mammalian cells. To form chromatin, DNA is tightly condensed by being wrapped around nuclear proteins called histones. This repeating DNA-histone complex, which consists of 146 base pairs of double-stranded DNA wrapped around eight histone proteins, is called a nucleosome. The repeating nucleosomal units are commonly referred to appear as “beads on a string” when seen under a microscope.
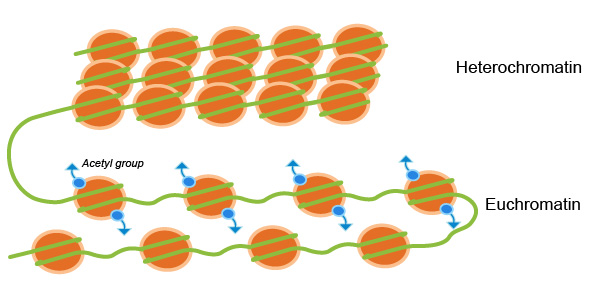
In general, the more condensed the chromatin, the harder it is for transcription factors and other DNA binding proteins to access DNA and perform their duties. When chromatin is tightly packed, and not actively being transcribed it is called heterochromatin. When chromatin is more loosely packed, and therefore accessible for transcription it is called euchromatin (Fig. 1). Chromatin remodeling is highly implicated in epigenetics. Epigenetic modifications to histone proteins such as methylation/demethylation and acetylation/deacetylation can alter the structure of chromatin resulting in transcriptional activation or repression.1
Tools of the Trade
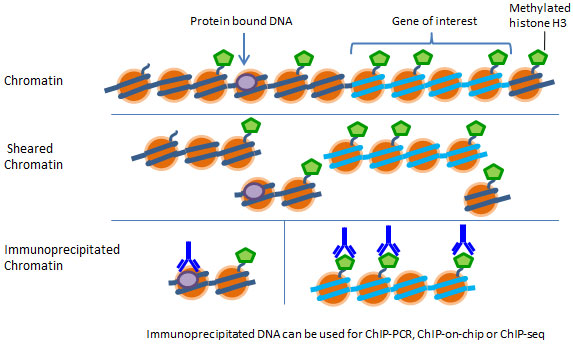
Chromatin Immunoprecipitation (ChIP)
Chromatin is the complex of DNA and proteins that are packed within the nucleus of eukaryotic cells. The interaction of these DNA and proteins is critical for cellular functions such as signal transduction, gene transcription, chromosome segregation, DNA replication and recombination, and epigenetic silencing.
Chromatin immunoprecipitation (ChIP) offers an advantageous tool for studying protein-DNA interactions. It can be used for determining if a specific protein binds to a particular sequence of a gene, such as the target sequence of a transcription factor or to compare the level of histone methylation associated with a specific gene promoter region between normal and diseased tissue. Identifying the genetic targets of DNA binding proteins and revealing the mechanism of protein-DNA interaction is crucial for understanding cellular processes.
For a ChIP assay, chromatin is extracted, sheared and then added into a microwell where protein-DNA complexes are captured by affinity antibodies. The captured DNA is then released and purified. Eluted DNA can be used for various downstream applications including ChIP-PCR, ChIP-on-chip, and ChIP-seq.
First, isolate chromatin from your samples of interest and shear your chromatin down to 300 to 1000 base pairs in length, then utilize a microplate-based ChIP method paired with ChIP-grade antibodies of interest for fast and easy chromatin immunoprecipitation. The chromatin can then be used in PCR, microarray, or sequencing applications.
A nucleosome consists of two copies of each of the core histone proteins H2A, H2B, H3 and H4. These histone proteins are involved in structural organization of chromatin in eukaryotic cells and can undergo various posttranslational modifications which can alter their interaction with DNA and nuclear proteins. The H3 and H4 histones for example, have long tails protruding from the nucleosome which can be covalently modified at several sites. Modifications of the histone tails include methylation, acetylation, phosphorylation, ubiquitination, sumoylation, citrullination, and ADP-ribosylation occuring on the N-terminal tail domain, which result in remodeling of the nucleosome structure into an open conformation more accessible to transcription complexes (H2A can also be modified).
Combinations of these modifications are thought to constitute the so-called “histone code” which can be read and interpreted by different cellular factors leading to transcriptional activation or repression.2 For example, a general characteristic of euchromatin is tri-methylation at lysine residues of histone H3 – lysine 4 (K4), lysine 36 (K36), and lysine 79 (K79) and a high level of histone acetylation, while heterochromatin is characteristically enriched in trimethylation of other histone H3 lysine residues K9, K20, and K27.
Ready to learn about another epigenetic mechanism? Read on: Histone Modifications