What is Epigenetics technical articles are geared towards epigenetic research techniques, news, and trends in the field of epigenetics, written by scientists from universities and institutions including UCLA, Hofstra, NIH, Johns Hopkins, and more.
Explore in detail new epigenetic research techniques and tips for topics like next-generation sequencing (NGS) in epigenetics, m6A RNA methylation, CRISPR/Cas9 system epigenetic editing, chromatin immunoprecipitation (ChIP) protocol optimization, and single-cell epigenomics methods.
Want to share your research? Submit your own epigenetics article to be featured.
The recent FDA approval of Donanemab has sparked celebration within the Alzheimer’s research community, offering significant hope by slowing cognitive decline in some patients. However, it is clear to scientists that Donanemab is not a cure, and the quest to truly halt or reverse Alzheimer’s disease (AD) remains ongoing. Donanemab works by targeting amyloid beta (Aβ) plaques, a hallmark protein buildup in AD’s pathology. While clearing these plaques marks a critical step, it may not address the root cause of [more…]
Understanding trait inheritance is crucial for unraveling the complexities of life’s blueprint and how it shapes the world around us. It provides insights into the transmission of characteristics from one generation to the next, shedding light on the mechanisms that underpin biological diversity and evolution. In a groundbreaking study featured in the journal Nature, researchers have unveiled the hidden secrets of how our DNA replication machinery meticulously safeguards and transfers epigenetic information during cell division. Led by Professor Yuanliang Zhai [more…]
In the quest to understand the intricate interplay between genetics and the environment in disease development, scientists have devoted extensive efforts to unraveling the structure and sequence of genetic material. A key focus of this research lies in exploring epigenetic marks, the chemical modifications made on DNA or its packaging proteins (histones) that affect gene expression without changing the DNA sequence. Epigenetic modifications serve as crucial regulators of gene activity, orchestrating when and how genes are turned on or off [more…]
This article was first published by EpigenTek. Since the 1970s when studies on the epigenetics of chromatin first started, histone and DNA marks, especially DNA methylation, have become the most investigated and characterized epigenetic modifications to date [Razin]. DNA methylation has garnered significant attention within the field as a pivotal regulator of gene expression. Its extensive influence on biological processes, ranging from development to disease, has redefined our comprehension of genetic control and prompted innovative approaches for medical interventions. During development, [more…]
This article was first published by EpigenTek. It would be a near impossible feat for the human genome, consisting of some 3 billion bases, to fit within its relatively miniscule cellular accommodations if not for the help of a family of very critical DNA-interacting proteins. Histones are the primary protein constituent of chromatin, forming complexes with DNA to compact our rather expansive genetic material for efficient nuclear organization. But make no mistake, these protein “spools” are not merely packaging units [more…]
Think of DNA as a thread wound around a spool. It’s packed and wrapped around structures called nucleosomes, resembling beads along the thread. This compact packaging provides organization and protection for the DNA. However, in areas where the DNA is densely packed, it can pose a challenge for proteins attempting to reach it. Interestingly, there are certain proteins that possess the remarkable ability to access tightly wound DNA. These proteins, known as pioneer transcription factors, have evolved special mechanisms that [more…]
All cancerous tumors exhibit these reversible epigenetic changes that could represent novel reprogrammable targets to address drug resistance (1). Recent research published in Communications Biology provides essential information about the specific epigenetic alterations that may be responsible for chemotherapy resistance (2). Drug Resistance in Cancer More than 90% of the mortality caused by cancer can be linked to drug resistance (3). The molecular mechanisms mediating drug resistance in cancerous cells include increased function of enzymes that render drugs ineffective, mutations [more…]
Associations between prenatal exposure to chemicals like endocrine disruptors found in plastics and the development of neurological diseases later in life have been well described (1). Although mechanisms have been proposed for how endocrine disruptors affect human health (2), those mediating the development of neurological conditions like autism and dementia have been difficult to elucidate because of the multifactorial nature of these diseases. Researchers, therefore, considered the issue through a new lens: inherited DNA methylation patterns. Bisphenols and phthalates are [more…]
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first recognized at the beginning of 2020 and is responsible for the ongoing COVID-19 pandemic. The persistence of the virus has been partly accredited to its effective suppression of host cell responses. As such, continued research in elucidating the dynamics of the SARS-CoV-2 life cycle is essential to facilitate the design and development of novel diagnostics and suitable therapies. Although reports suggest that SARS-CoV-2 can dysregulate the host’s gene expression and innate [more…]
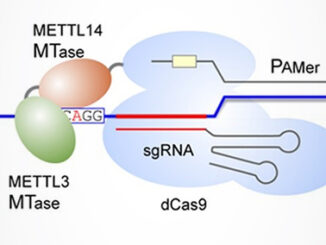
“The fifth RNA base” N6-methyladenosine, or m6A, is the most common and abundant eukaryotic RNA modification, accounting for over 80% of all RNA methylation. It can be found mainly in mRNA, but is also observed in non-coding species like tRNA, rRNA, and miRNA. Through interactions with various binding proteins called “readers”, m6A affects virtually every facet of ribonucleic acid biology: structure, splicing, localization, translation, stability, and turnover [1]. Aside from this central role in RNA metabolism, m6A is a factor [more…]