“The fifth RNA base”
N6-methyladenosine, or m6A, is the most common and abundant eukaryotic RNA modification, accounting for over 80% of all RNA methylation. It can be found mainly in mRNA, but is also observed in non-coding species like tRNA, rRNA, and miRNA. Through interactions with various binding proteins called “readers”, m6A affects virtually every facet of ribonucleic acid biology: structure, splicing, localization, translation, stability, and turnover [1]. Aside from this central role in RNA metabolism, m6A is a factor in other physiological processes such as cell differentiation, immunity, inflammation, and the circadian clock [2].
Mechanism of RNA m6A addition and deletion
The formation of m6A RNA appears to be a co-transcriptional event taking place early on in the RNA lifecycle and is mediated by a multi-protein methyltransferase complex composed, in part, of METTL enzymes (Figure 1). Chemical addition of a methyl group at the nitrogen-6 position of adenosine (A) residues to form m6A occurs via the METTL3/14 heterodimer. FTO-mediated oxidative demethylation converts m6A in a step-wise manner to hm6A and subsequently f6A before finally reverting back to A.
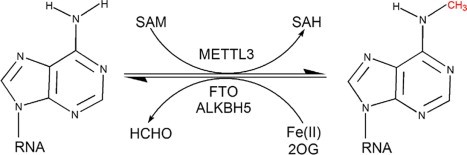
The recent discoveries of these methylase “writers” (METTL3/14, WTAP, RBM15/15B, KIAA1429) and their associated demethylase “erasers” (FTO, ALKBH5, TET-like enzymes) in mammals uncovered the reversibility of the m6A modification, revealing potential therapeutic targets for m6A dysregulation-related diseases. Abnormal m6A methylation has been implicated in diverse pathologies: diabetes, obesity, neurodegeneration, and cancer, to name a few.
Applying CRISPR-Cas to the epitranscriptome
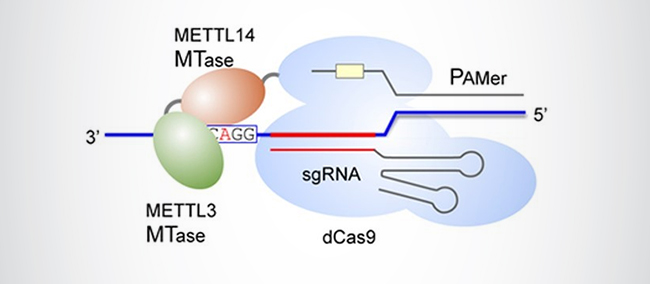
Since its role in prokaryotes as an adaptive immune system against invading DNA bacteriophages was elucidated [4], CRISPR-Cas soon made its way into the realm of genetic engineering, being fashioned into a programmable gene-editing tool capable of artificially modifying an organism’s genome by precisely cleaving dsDNA sequences at desired loci [5]. The system in this context consisted of two main components, a synthetic guide RNA complexed with the Cas9 endonuclease for site-specific recognition and cleavage, respectively, of the host DNA target.
Shortly thereafter, attention was turned toward RNA substrates. Noting that Cas9 functionality is contingent upon the nuclease’s recognition of a short genomic DNA sequence known as the protospacer-adjacent motif (PAM), which RNA lacks, site-specific Cas9-mediated cleavage of ssRNA targets was achieved by the inclusion of a third component, a separate PAM-containing DNA oligonucleotide (or PAMmer) [6].
The advent of m6A editing systems

The ability to customize these components has afforded CRISPR-Cas with limitless potential in basic research, biotechnology, and medicine. More recently, it has found utility in epigenetic-based applications. In 2019, researchers developed CRISPR-Cas systems targeting the epitranscriptome, a method dubbed “m6A editing” [7]. By coupling RNA-guided dead Cas9 (dCas9; catalytically inactive Cas9 that can serve as a delivery vehicle for other enzymes of interest) with either a single-chain m6A methyltransferase (a fusion protein composed of METTL3- and METTL4-derived MTase domains; Figure 2) or full-length demethylases, a series of precision m6A editors (M3M14-dCas9, ALKBH5-dCas9, and FTO-dCas9) was generated with the capacity to manipulate mRNA methylation at specified transcript locations, without affecting the underlying primary RNA sequence.
Refining m6A editors
Subsequent refinements to m6A editing involved substituting dCas9 for dCas13b [8,9], an endonuclease that, in its active form, specifically targets and cleaves ssRNA. Cas13’s innate preference for RNA precludes the need for an additional PAMmer component required by Cas9 m6A editors. In addition, while M3M14-dCas9 was found to primarily localize in the cytoplasm, dCas13b constructs were tailored for either cytosolic or nuclear localization via tagging with nuclear export signal (NES) and nuclear localization signal (NLS) sequences, respectively, allowing for wider access to m6A reader proteins [9]. Similarly, NES-tagged ALKBH5-dCas13b could be detected in both the cytoplasm and nucleus [8], in contrast to nuclear-localized ALKBH5-dCas9.
Another Cas13 ortholog, RfxCas13d (aka CasRx), has been investigated in epitranscriptome editing as its smaller size favors more efficient packaging into viral vectors for delivery into hard-to-transfect cells, with NLS-tagged METTL3-dCasRx and ALKBH5-dCasRx conjugates displaying site-specific m6A writer and eraser activity, respectively [10].
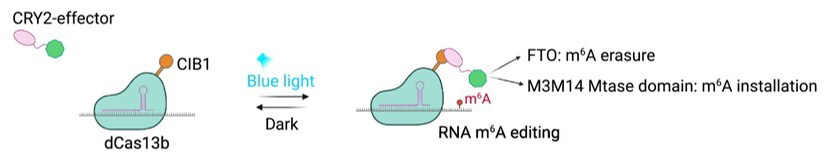
While these systems can regulate m6A modification spatially, an innovative approach involving light-inducible protein-protein interactions permits both spatial and temporal regulation of m6A editing (Figure 3). CRY2 is a photosensitive protein that binds the CIB1 protein upon stimulation with blue light. By tethering each protein to a different component of a CRISPR-Cas m6A RNA editing system (CIB1 fused to dCas13b and CRY2 fused to m6A effectors), CIB1-CRY2 dimerization can be optically controlled to form a functional complex for m6A installation or erasure [11].
Aside from m6A writers and erasers, programmable Cas-m6A reader proteins have also been engineered to study RNA regulation at the single RNA level. Fusions of the reader proteins YTHDF2 and YTHDF1 with RNA-guided dCas13b were fashioned whereby degradation and enhanced translation, respectively, of target mRNA transcripts could be site-specifically induced [13].
Equip your research
Ongoing and future research in m6A editing will undoubtedly benefit from reliable assays for the detection of the m6A RNA modification and its modifying enzymes. Our sponsor EpigenTek offers a comprehensive suite of products to facilitate your RNA methylation research needs, including RNA extraction, global m6A RNA quantification, m6A methylase and demethylase activity/inhibition assessment, m6A RNA enrichment, and m6A antibodies.
References
- Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20(10):608-624. doi:10.1038/s41580-019-0168-5
- Hastings MH. m(6)A mRNA methylation: a new circadian pacesetter. Cell. 2013;155(4):740-741. doi:10.1016/j.cell.2013.10.028
- Niu Y, Zhao X, Wu YS, Li MM, Wang XJ, Yang YG. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteomics Bioinformatics. 2013;11(1):8-17. doi:10.1016/j.gpb.2012.12.002
- Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709-1712. doi:10.1126/science.1138140
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816-821. doi:10.1126/science.1225829
- O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516(7530):263-266. doi:10.1038/nature13769
- Liu XM, Zhou J, Mao Y, Ji Q, Qian SB. Programmable RNA N6-methyladenosine editing by CRISPR-Cas9 conjugates. Nat Chem Biol. 2019;15(9):865-871. doi:10.1038/s41589-019-0327-1
- Li J, Chen Z, Chen F, et al. Targeted mRNA demethylation using an engineered dCas13b-ALKBH5 fusion protein. Nucleic Acids Res. 2020;48(10):5684-5694. doi:10.1093/nar/gkaa269
- Wilson C, Chen PJ, Miao Z, Liu DR. Programmable m6A modification of cellular RNAs with a Cas13-directed methyltransferase. Nat Biotechnol. 2020;38(12):1431-1440. doi:10.1038/s41587-020-0572-6
- Xia Z, Tang M, Ma J, et al. Epitranscriptomic editing of the RNA N6-methyladenosine modification by dCasRx conjugated methyltransferase and demethylase. Nucleic Acids Res. 2021;49(13):7361-7374. doi:10.1093/nar/gkab517
- Zhao J, Li B, Ma J, Jin W, Ma X. Photoactivatable RNA N6 -Methyladenosine Editing with CRISPR-Cas13. Small. 2020;16(30):e1907301. doi:10.1002/smll.201907301
- Lan TH, He L, Huang Y, Zhou Y. Optogenetics for transcriptional programming and genetic engineering [published online ahead of print, 2022 Jun 20]. Trends Genet. 2022;S0168-9525(22)00140-8. doi:10.1016/j.tig.2022.05.014
- Rauch S, He C, Dickinson BC. Targeted m6A Reader Proteins To Study Epitranscriptomic Regulation of Single RNAs. J Am Chem Soc. 2018;140(38):11974-11981. doi:10.1021/jacs.8b05012
