
This article was first published by EpigenTek.
Since the 1970s when studies on the epigenetics of chromatin first started, histone and DNA marks, especially DNA methylation, have become the most investigated and characterized epigenetic modifications to date [Razin]. DNA methylation has garnered significant attention within the field as a pivotal regulator of gene expression. Its extensive influence on biological processes, ranging from development to disease, has redefined our comprehension of genetic control and prompted innovative approaches for medical interventions.
During development, DNA methylation patterns shape the differentiation of cells, ensuring the emergence of distinct cell types with specialized functions. Aberrancies in these patterns have been implicated with cancer and other maladies. For instance, hypermethylation of tumor suppressor gene promoters and hypomethylation of oncogene promoters contribute to uncontrolled cell growth, fueling cancer progression. As investigators probe deeper into the complexities of DNA methylation, new avenues for understanding and potentially modulating this epigenetic mark are being opened, offering transformative advancements in biomedical research and healthcare.
Editing the epigenome
Among these revolutionary innovations, epigenome editing has been making quite an impact. This method involves the targeted modification of epigenetic marks, such as DNA methylation, RNA methylation, and histone modifications, to regulate gene expression without altering the underlying primary DNA sequence. With regard to DNA methylation, epigenome editing focuses on changing the pattern of methyl groups attached to cytosine bases in DNA. One common way to achieve this is via engineered molecules or proteins like DNA methyltransferases (DNMTs) to add methyl groups or demethylases to remove them in a targeted manner, and the CRISPR-Cas system is often tailored for this purpose.
Design principles and strategies
Since its role in prokaryotes as an adaptive immune system against invading DNA bacteriophages was elucidated [Barrangou], CRISPR-Cas soon made its way into the realm of genetic engineering, being fashioned into a programmable gene-editing tool capable of artificially modifying an organism’s genome by precisely cleaving dsDNA sequences at desired loci [Jinek]. The system in this context consisted of two main components: a synthetic guide RNA complexed with the Cas9 endonuclease for site-specific recognition and cleavage, respectively, of the host DNA target.
The ability to customize these components has afforded CRISPR-Cas with limitless possibilities in basic research, biotechnology, and medicine. More recently, it has found utility in epigenetic-based applications. In 2016, several research groups independently developed CRISPR-Cas systems targeting the epigenome [Choudhury, Liu, McDonald, Vojta]. By fusing RNA-guided dead Cas9 (dCas9; catalytically inactive Cas9 that can serve as a delivery vehicle for other enzymes of interest) with either a 5mC methyltransferase (dCas9-Dnmt3a) or demethylase (dCas9-Tet1), a series of precision 5mC editors was generated with the capacity to manipulate DNA methylation at specified genomic loci.
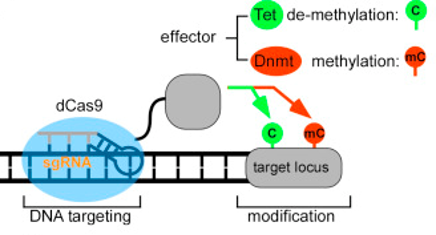
Schematic representation of CRISPR-Cas-based DNA methylation editors (from Figure 1A, [Liu])
As Dr. Boris Kantor, Professor of Neurobiology and Director of the Viral Vector Core at Duke University, details, “The CRISPR-Cas used for epigenome-editing within a locus-of-interest consists of several components. dCas9 as mentioned above is linked to the DNA methylase repressor usually at the C-terminus. To transport the complex into the nucleus, two or even four nuclear localization signals (NLSs) are used at the N- and C-terminus of the complex. dCas9 and DNA methylase are usually separated via a short polypeptide linker supporting a proper folding of the polypeptides.”
“Target-specific DNA methylation modifications rely on Cas9 fused to TET (to demethylate) or DNMT3A (to methylate) domains,” states Dr. Alba Rodriguez-Meira, Sir Henry Wellcome Postdoctoral Fellow at the Dana-Farber Cancer Institute and the Broad Institute of MIT and Harvard. Dr. Daniel Sapozhnikov, Scientist I at AvenCell Therapeutics, further elaborates, “If the cells are not dividing, … some version of a TET enzyme – which is the closest thing we currently have to a ‘demethylase’ – would need to be used as part of a dCas9 fusion. For methylation, there are now numerous variations and iterations of fusions or recruitment of methyltransferases by dCas9.”
Applications
Epigenome editing has various applications, including basic science research and prospective therapeutic interventions for diseases like cancer and genetic disorders.
“I would say the two main categories with the highest chance to move into the clinical space would be rare diseases and cancer,” proposes Dr. Kantor. “For example, imprinting diseases, such as Prader-Willi and Angelman syndromes, are rare congenital disorders caused by aberrant dosages of imprinted genes and are great candidates for methylation-based editing.
“Disruption of genetic or epigenetic mechanisms, specifically DNA methylation, can lead to issues with regulating the expression of imprinted genes, thus causing diseases. Catalytically dead Cas9 tethered with DNA de- and methyltransferases have been shown to have high epigenome editing efficiency in correcting those diseases (predominately in vitro).
“Another disease which would likely be a good target for CRISPRi editing is Rett syndrome. Last, but not least, the CNS diseases could be efficiently targeted via DNA methylation. In fact, we demonstrated that.” Dr. Kantor is referring to work that he, along with Dr. Ornit Chiba-Falek, Professor of Neurology and Division Chief of Translational Brain Sciences at Duke University, and colleagues, published discussing the role of elevated SNCA levels in Parkinson’s disease (PD) and proposing a therapeutic strategy involving targeted DNA methylation within SNCA intron 1 using a CRISPR-deactivated Cas9 system [Kantor]. The developed system, a lentiviral vector carrying dCas9 and DNMT3A, was shown to successfully downregulate SNCA expression in human-induced pluripotent stem cell-derived dopaminergic neurons, offering a potential novel epigenetic-based therapeutic approach for PD.
Actual clinical application of CRISPR-Cas-based DNA methylation editors, though, is a work in progress. “To the best of my knowledge,” says Dr. Rodriguez-Meira, “all clinical trials trying to modulate DNA methylation in cancer rely on broad DNA methyltransferase inhibitors (azacytidine and derivatives) and a novel small molecule DNMT1 inhibitor in the context of acute myeloid leukemia [Pappalardi]. However, epigenomic editing tools are still far away of being deployed into the clinic, and to my knowledge none of them are in clinical trials.”
“There are not yet any clinical trials using DNA methylation editing technology,” Dr. Sapozhnikov corroborates, although the biotechnology sector has shown progress in improving this outcome. “Start-ups have reported successes in animal models using epigenetic editing technology (Tune Therapeutics, but also Chroma Medicine being the most developed one in the DNA methylation editing space) … Tune Therapeutics is using epigenetic editing technology (not DNA methylation editing, but of similar principle) like this to turn on or off genes that enhance the function of T-cells that were engineered to fight cancer (CAR-T cells), rather than to change the cancer cells themselves. In this way, epigenetic editing can be used to enhance this cell-based anticancer therapy.”
Key challenges and limitations
DNA methylation editing, as with any early-stage technology, has its fair share of shortcomings necessitating fine-tuning. According to Dr. Kantor, “There are two main limitations of the methylation-based editors: size and efficiency. The size of the tools based on the DNA methylation effectors is large and it is prohibitive for efficient packaging and delivery with adeno-associated vectors [AAVs].
“The second limitation is that the efficiency of those tools is relatively low. This stems from two reasons. First, DNA methylation controls gene expression in limited genetic loci, and even in those regions is not the sole mechanism mediating the repressive process. Second, de novo methyltransferase A and B many times need an additional enzyme DNMT3L which acts as a chaperon that enhances its activity. As such, multipartite sequences are needed to get reliable and sustainable effects.”
Dr. Rodriguez-Meira concurs, “Key challenges are the big size of Cas9-based epigenomic editors, which makes it really difficult to use these tools in primary cells. Achieving CpG resolution with these tools is extremely challenging (i.e., to target only one specific CpG and not other CpGs around it) because Cas9-TET and Cas9-DNMT3A are big proteins.
“Therefore, at the practical level, these tools bring TET and DNMT3A in close proximity to a big target region (300-500 bp), which gets methylated or demethylated in bulk. In addition to this challenge, the efficiency of DNA methylation editors is still extremely low, requiring the use of several guide RNAs ‘tiling’ the target regions to achieve sufficient signal. Therefore, high-efficiency editing is typically restricted to sites that have high density of guide RNA designs around them and high CpG content.”
Off-target effects are another drawback that still requires attention. “These enzymes can also work pretty well throughout the genome, even if they are tied down to and targeted by a bulky dCas9,” Dr. Sapozhnikov points out. “This creates unintended off-target changes that are widespread throughout the genome and may negatively impact both research and therapy by confounding the experimental question or by causing deleterious changes, respectively.”
Dr. Sapozhnikov continues, “Also, will the changes introduced by these techniques be stable? Will the effect of the DNA methylation change be enough to dramatically change gene expression, and then, will that be enough to alter the course of the disease? There are many questions that must be answered and many challenges to be resolved.”
Addressing the issues
Researchers are continually developing new tools and techniques to improve the precision and efficiency of epigenome editing.
“First, DNA methyltransferases can be packaged into other vector systems, e.g., lentiviral vector,” Dr. Kantor says. While AAVs are a commonly used delivery platform due to their versatility in transducing both dividing and nondividing cells, as well as their low immunogenicity, their limited cargo capacity can restrict the size of the payload they can carry. In response, Dr. Kantor and coworkers developed an integrase-deficient lentiviral vector for CRISPR/Cas9 delivery with low host chromosome integration capability, similar to AAVs [Ortinski]. “The packaging capacity of this system is twice larger than that of the AAV.
“Second, more sophisticated design of gRNA to the methylation ‘sensitive’ loci could help to improve its efficiency. As we demonstrated, the methylation can be brought to the region that is far away from the site of interest, and still be very efficient in silencing of the desired site. Accordingly, we and others suggested that the spread of DNA methylation could reach regions which are mega base pairs away from the desired DNA loci.”
On the other hand, Dr. Sapozhnikov developed a unique epigenetic engineering technique that does not involve the use of an enzyme fusion. Rather, it relies on the steric hindrance of bound dCas9 to physically impede the endogenous DNA methylation machinery at target sites, thereby attenuating undesirable enzyme-associated side effects [Sapozhnikov].
You May Also Want to Read:
- Nuclear Extraction Protocol
- Comparison of Methods for Quantification of Global DNA Methylation
- Abnormal DNA Methylation Induced by TET Repression
As Dr. Sapozhnikov explains, “These techniques typically rely on fusions between enzymes that edit DNA methylation and a nuclease-dead Cas9. These are not ‘clean’ enzymes that modify only DNA methylation. They have evolved over millions of years to participate in a coordinated epigenetic network – they interact with other proteins to cause many changes besides just methylation…. The tool we developed is, in my opinion, the best tool for DNA demethylation editing in dividing cells, because it does so cleanly and efficiently.”
Regarding other current breakthroughs in the field, “In my view, some of the most exciting discoveries in DNA methylation editing relate to novel tools that describe how these edits can be stably propagated over time and impart ‘epigenetic memory’, including transgenerational inheritance,” submits Dr. Rodriguez-Meira. “For example, articles like [the recent paper] from Juan Carlos Izpisua Belmonte,” in which evidence was presented supporting transgenerational epigenetic inheritance in mammals (specifically in mice) by demonstrating the transmission of DNA methylation of promoter-associated CpG islands (CGIs) from parents to offspring [Takahashi]. The study utilized DNA methylation-edited mouse embryonic stem cells, showing that targeted CGI methylation and associated abnormal metabolic phenotypes were maintained and transmitted across multiple generations.
Dr. Sapozhnikov shares a similar sentiment. “Perhaps the most interesting recent discovery has nothing to do with the development of the technology (which, I feel, has slowed, due to the limited number of enzymes and combinations thereof that can be used in the technology) but with the application of the technology. Gabriella Ficz’s group recently published a paper [Saunderson] that showed that DNA methylation changes that are introduced by editing can be maintained as blood cells divide and differentiate (i.e., inherited by new cells), meaning that changes in DNA methylation intended to cure disease could be long-lasting.
“This suggests that, despite the fact that DNA methylation changes are relatively easily reversible (unlike changes to the DNA sequence), they could still be permanent (or at least stable), allowing for one-and-done or low-frequency treatments. Of course, from a mechanistical standpoint, we must again ask if other non-DNA-methylation-related epigenetic effects caused by this technology (DNMT3A recruitment of other factors) are contributing to this inheritance, but therapeutically this is fantastic news!”
DNA methylation editing holds great promise for advancing our knowledge of gene regulation and developing new strategies for addressing genetic and epigenetic disorders. Researchers in the field continue to make significant strides, and the technology is likely to have a growing impact on biology and medicine.
References
- Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709-1712. doi:10.1126/science.1138140
- Choudhury SR, Cui Y, Lubecka K, Stefanska B, Irudayaraj J. CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget. 2016;7(29):46545-46556. doi:10.18632/oncotarget.10234
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816-821. doi:10.1126/science.1225829
- Kantor B, Tagliafierro L, Gu J, et al. Downregulation of SNCA Expression by Targeted Editing of DNA Methylation: A Potential Strategy for Precision Therapy in PD. Mol Ther. 2018;26(11):2638-2649. doi:10.1016/j.ymthe.2018.08.019
- Liu XS, Wu H, Ji X, et al. Editing DNA Methylation in the Mammalian Genome. Cell. 2016;167(1):233-247.e17. doi:10.1016/j.cell.2016.08.056
- McDonald JI, Celik H, Rois LE, et al. Reprogrammable CRISPR/Cas9-based system for inducing site-specific DNA methylation. Biol Open. 2016;5(6):866-874. Published 2016 Jun 15. doi:10.1242/bio.019067
- Ortinski PI, O’Donovan B, Dong X, Kantor B. Integrase-Deficient Lentiviral Vector as an All-in-One Platform for Highly Efficient CRISPR/Cas9-Mediated Gene Editing. Mol Ther Methods Clin Dev. 2017;5:153-164. Published 2017 Apr 19. doi:10.1016/j.omtm.2017.04.002
- Pappalardi MB, Keenan K, Cockerill M, et al. Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia. Nat Cancer. 2021;2(10):1002-1017.
- Sapozhnikov DM, Szyf M. Unraveling the functional role of DNA demethylation at specific promoters by targeted steric blockage of DNA methyltransferase with CRISPR/dCas9. Nat Commun. 2021;12(1):5711. Published 2021 Sep 29. doi:10.1038/s41467-021-25991-9
- Saunderson EA, Encabo HH, Devis J, et al. CRISPR/dCas9 DNA methylation editing is heritable during human hematopoiesis and shapes immune progeny. Proc Natl Acad Sci U S A. 2023;120(34):e2300224120. doi:10.1073/pnas.2300224120
- Takahashi Y, Morales Valencia M, Yu Y, et al. Transgenerational inheritance of acquired epigenetic signatures at CpG islands in mice. Cell. 2023;186(4):715-731.e19. doi:10.1016/j.cell.2022.12.047
- Vojta A, Dobrinić P, Tadić V, et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016;44(12):5615-5628. doi:10.1093/nar/gkw159

