Post-translational modifications are well known for their influence on protein stability, enzymatic functions, as well as protein:protein interactions. At the level of gene expression, acetylation, methylation, phosphorylation, ADP-ribosylation, ubiquitination and SUMOylation are some of the most prevalent chemical modifications that regulate transcription factors and the chromatin template (the complex of DNA and histone proteins) alike. In particular, the manifold modifications occurring on the histone proteins that closely associate with DNA are proposed to constitute a gene-regulating code (the histone code). That is, combinations of specific modifications distinguish active (gene switched on) or silent (gene switched off) states of chromatin, and are thought to control how tightly the DNA wraps around histones as well as provide ‘docking sites’ for factors that remodel chromatin.
Because these modifications can regulate changes in gene expression independently of the DNA sequence, they are described as epigenetic. Adding a further layer of complexity, many epigenetic enzymes modify a broad range of histone and non-histone proteins. For instance, the Set7 enzyme methylates lysine residues on H3 histones as well as numerous transcription factors and other epigenetic enzymes, with varying effects on gene expression.
Emerging evidence indicates that post-translational modification of histones and transcription factors link local environmental changes with gene expression. Moreover, this intricate signaling network is increasingly implicated in sensing metabolic fluctuations and changes in cellular energy state.
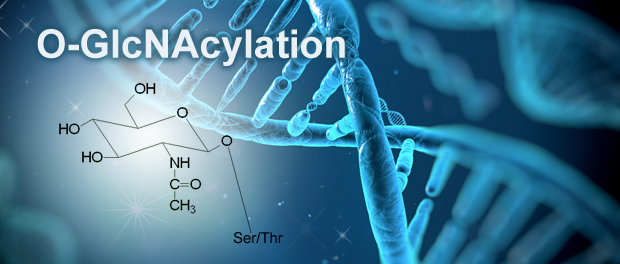
The nutrient-responsive hexosamine biosynthetic pathway (HBP) is acutely sensitive to carbohydrate, lipid, amino acid, and ATP levels. Activation of the HBP leads to increased synthesis of UDP-GlcNAc, a key metabolite in the dynamic O-linked β-N-acetylglucosamine (O-GlcNAc, pronounced Oh-Glick-Nack) modification at serine and threonine residues of a growing list of cytoplasmic and nuclear proteins essential for cell survival. A pair of highly conserved enzymes dynamically regulates O-GlcNAcylation; the O-GlcNAc transferase (OGT) adds the modification and the O-GlcNAcase (OGA) removes it. Interestingly, both high glucose and glucose deprivation have been shown to increase protein O-GlcNAcylation. Furthermore, the recent discovery that O-GlcNAcylation activates the AMPK monitor of cellular energy levels highlights a crucial role for this post-translational modification in metabolism.
In addition, studies have begun to define a role for O-GlcNAcylation in human disease. Proteins involved in vascular function, calcium handling, and the adverse effects of diabetes on the heart are modified by O-GlcNAcylation. Intriguingly, O-GlcNAc-mediated activation of a key epigenetic enzyme important for cardiac function, the Ezh2 lysine methylase, potentially links the HBP with chromatin modifications.
In fact a classical epigenetic role for this modification has recently emerged from the identification of distinct sites for O-GlcNAcylation on each of the four core histone proteins (Table 1). While studies are yet to completely define the function of O-GlcNAc on histones, several of these newly identified sites are also known locations for other modifications. For example, O-GlcNAcylation of H3 histones at threonine-32 competitively regulates phosphorylation of the same residue to control cell cycle progression. Similarly, GlcNAcylation facilitates the ubiquitination of serine 112 on histone H2B, and is frequently found near genes that are switched on. In addition, OGA is also able to catalyze the gene-activating acetyl modification on histones. It is therefore reasonable to speculate that OGA could couple the removal of O-GlcNAc while increasing histone acetylation.
Until recently, methylation at the 5-carbon ring of cytosine nucleotides classically associated with gene silencing, was the only epigenetic mark known to occur on DNA. Recent experimental evidence implicates O-GlcNAc in regulating the subcellular localization and enzymatic activity of the Ten Eleven Translocation (TET) dioxyegnases that are implicated in transitions between methylated and unmethylated DNA. Unexpectedly, TETs were shown to regulate OGT chromatin localization.
Indeed O-GlcNAcylation can epigenetically influence gene expression by several distinct, yet interconnected mechanisms. Newly developed quantification techniques allowing genome-wide profiling of histone O-GlcNAcylation will greatly enhance our understanding of chromatin regulation. Likewise, proteomic studies are likely to reveal further influences of O-GlcNAc on the enzymatic machinery that catalyze the addition and removal of histone and DNA modifications. Characterization of the O-GlcNAc – epigenetic connection has strong potential to identify important functions in metabolism as well as human disease.
Table 1. Chromatin targets of GlcNAc
| Histone | Modification site | Specific function |
| H2A | threonine 101 | Unknown |
| H2B | serine 36 | May compete with serine 36 phosphorylation |
| H2B | serine 112 | Promotes H2B-K120 monoubiquitination, presumably for transcriptional activation |
| H3 | threonine 32 | Competes with mitosis-specific phosphorylation of H3-serine 10 and H3- serine 28 |
| H3 | serine 10 | Suppress phosphorylation of serine 10 |
| H4 | serine 47 | Unknown |
References
- Jenuwein T, Allis CD. (2001). Translating the histone code. Science, 293(5532):1074-80
- Keating ST, El-Osta A. (2013). Transcriptional regulation by the Set7 lysine methyltransferase. Epigenetics, 8(4):361-72
- Kaelin WG, McKnight SL. (2013). Influence of metabolism on epigenetics and disease. Cell, 153(1):56-69
- Hart GW. (2014). Three decades of research on GlcNAcylation – a major nutrient sensor that regulates signaling, transcription and cellular metabolism. Front Endocrinol, 5:183
- Zou L, et al., (2012). Glucose deprivation-induced oncrease in protein O-GlcNAcylation in cardiomyocytes is calcium-dependent. J Biol Chem, 287(41):34419-31
- Xu Q, et al., (2014). AMPK regulates histone H2B O-GlcNAcylation. Nucleic Acids Res, 42(9):5594-604
- Hilgers RH, et al., (2012). Acute O-GlcNAcylation prevents inflammation-induced vascular dysfunction. Am J Physiol Heart Circ Physiol, 303(5):H513-22
- Zhu-Mauldin X, et al., (2012). Modification of STIM1 by O-linked N-acetylglucosamine (O-GlcNAc) attenuates store-operated calcium entry in neonatal cardiomyocytes. J Biol Chem, 287(46):39094-106
- Marsh SA, et al., (2013). Cardian O-GlcNAcylation blunts autophagic signaling in the diabetic heart. Life Sci, 92(11):648-56
- Chu CS, et al., (2014). O-GlcNAcylation regulates EZH2 protein stability and function. PNAS, 111(4):1355-60
- Zhang S, et al., (2011). Modification of histones by sugar β-N-acetylglucosamine (GlcNAc) occurs on multiple residues, including histone H3 serine 10, and is cell cycle-regulated. J Biol Chem, 286(43):37483-95
- Fong JJ, et al., (2012). β-N-Acetylglucosamine (O-GlcNAc) is a novel regulator of mitosis-specific phosphorylations on histone H3. J Biol Chem, 287(15):12195-203
- Fujiki R, et al., (2011). GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature, 480(3778):557-60
- Toleman C, et al., (2004). Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J Biol Chem, 279(51):53665-73
- Zhang Q, et al., (2014). Differential regulation of the ten-eleven translocation (TET) family of dioxygenases by O-linked β-N-acetylglucosamine transferase (OGT). J Biol Chem, 289(9):5986-96
- Cehn Q, et al., (2013). TET2 promotes histone O-GlcNAcylation during gene transcription.


