
DNA methylation is an essential epigenetic mechanism that plays a critical role in the regulation of gene expression during embryonic development. The erasure of Tet-oxidized 5-Methylcytosine (5-mC) is not only important during the reprogramming steps but as well as the cell differentiation stages of development. In their study, Kweon et al. (2017) have shown that the SOS response-associated peptidase domain protein, SRAP1, binds to Tet-oxidized forms of 5-mC in DNA and selectively cleave DNA containing Tet-oxidized 5-methylcytosine. They also reveal a mechanism for targeted erasure of DNA cytosine methylation through the enzymatic actions of Tet and SRAP.
SRAP1 is an ortholog of the SRAP domain superfamily from the bacteria phyla which plays a role in DNA damage repair. Previous work from the Feldman lab revealed C3ORF37 may be part of the SRAP family and possess possible autoproteolytic activity. In this study, they show that C3ORF37/Hmces (Srap1) catalyzes the removal of 5mC, 5fC and 5caC. In order to show that SOS response-associated peptidase domain protein binds to Tet-oxidized forms of 5-mC, a DNA pull-down assay was performed using recombinant GST-Srap1 fusion protein. The recombinant protein stably bound to CpG sites containing 5hmC, 5fC, or 5caC but not 5mC in mouse ESCs (mESCs) extracts.
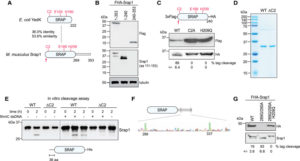
Due to the involvement of prokaryotic SRAP orthologs in the SOS response and its association in endo and exonuclease repair of DNA damage, Kweon et al. (2017) hypothesized that SRAP may have a possible nuclease function. In order to test this hypothesis, PCR products fully substituted with 5mC, 5hmC, 5fC, 5caC or unmodified cytosine were incubated with purified GST-Srap1. When ran out on a gel, modified DNA containing 5hmC, 5fC, or 5caC were degraded while DNA containing 5mC or unmodified cytosine were intact. Runoff-sequencing was then performed on partially digested DNA substrate containing 5hmC in place of cytosine and revealed cleavages immediately at 3’ of 5hmC and 5’ of major cut sites at CpG. These finding suggest that SRAP domain can catalyzes nucleolytic cleavage of DNA across sites containing Tet-oxidized 5mC.
CRISPR/Cas9 was then used to cause a deletion in the first coding exon of Srap1 in mouse zygote. Srap1 KO cells were not only morphologically different than WT cells but nuclear and perinuclear Srap1 immunoreactivity was detected in WT cell but not Srap1 KO cells. UHPLC-MS/MS analysis was performed on gDNA hydrolysates from WT and Srap1 KO cells which revealed an increase in 5fC, 5caC 5hmC and 5mC in Srap1 KO cells. However, when Srap1 was reintroduced to the KO cells the levels of 5mC and its Tet-oxidized derivatives were restored to levels comparable to WT ESCs.
To further examine the impact of Srap1 deficiency, RRBS was performed and further confirmed that Srap1 KO embryos exhibited a global increase in 5mC and 5hmC levels when compared to WT. The data from the CRISPR/Cas9-mediated gene targeting and RRBS studies revealed not only the function of Srap1 during embryogenesis but the changes caused by Srap1 deficiency.
Kweon et al. (2017) used DIP-seq to compare the distribution of Tet-oxidized 5mC derivatives between WT and Srap1 KO ESCs. Just as in the UHPLC analysis, DIP-seq resulted in an increase in the number of called 5hmC, 5fC and 5caC enriched regions in Srap1 KO cells when compared to WT. RNA-seq analysis showed different expression levels of certain genes in Srap1 KO cells, 98 genes were identified as being upregulated while 126 were downregulated.
Gene ontology analysis demonstrated that the genes that were downregulated were involved in the development and cell differentiation process. This finding was further confirmed by qPCR and the observed phonotypical changes in Srap1 deficient embryos. Several of the Srap1+/- embryos failed to develop properly resulting in masses of highly vascularized, undifferentiated tissues which is consistent with the downregulation of genes involved in the development and cell fate specification process.
The results of this study provide invaluable knowledge on the function of the previously uncharacterized Srap1 protein and an insight into the enzymatic mechanism of SRAP and Tet. The unique properties of the SRAP superfamily role in DNA damage repair is exploited in this mechanism for the erasure of Tet-oxidized 5-Methylcyotsine. SRAP1 provide an additional point of control that is used for the targeted removal of DNA cytosine methylation. This control may be critical for the reprogramming upon fertilization and during development.
Source: Kweon et al., Erasure of Tet-Oxidized 5-Methylcytosine by a SRAP Nuclease. Cell Reports, 2017. 21, p. 482-494.

