
It is a challenge to determine a cause and effect relationship between a distinct epigenetic mark and the ultimate behavior and function of the affected cell. However, CRISPR/Cas9 has provided the opportunity for investigators to manipulate the epigenome and observe the effects that it may have on cell function, development, and differentiation. The CRISPR/Cas9 system has been further expanded with the engineering of the nuclease-deficient version, dCas9, which can be used to directly manipulate a specific regulatory region or epigenetic marks to determine the impact on transcriptional activity and cellular functions.
Identification of cis-Regulatory Regions Using Cas9
Identifying functional regulatory regions, particularly distal enhancers, is a difficult task. Current methods for determining enhancer regions tend to have very large false discover rates. However, the CRISPR/Cas9 system can be used to determine enhancer regions that are associated with a gene of interest. CRISPR/Cas9 can be used to target putative non-coding regulatory regions and prevent protein binding and DNA interactions. gRNAs can be designed to introduce mutations within these regions, thereby changing the transcription of the associated gene. This method can confirm putative regulatory regions without having large false discovery rates. The only drawback to this approach is that in order for an investigator to study large genomic regions, they need to design a large number of gRNAs. However, the dCas9-based tool is very effective in inducing chromatin changes and it could reduce the number of gRNAs necessary.
The identification of regulatory regions can provide targets for epigenetic engineering resulting in the manipulation of gene expression or a better understanding of mechanisms underlying transcriptional regulation. Ultimately, this system can help validate complex DNA regulatory regions that may affect gene expression.
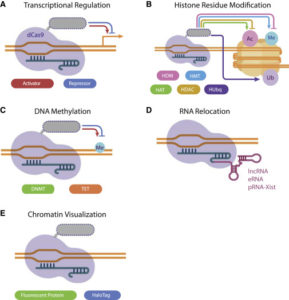
The dCas9-based tool may also be fused with a known functional domain, such as KRAB or SID, in order to repress gene transcription. On the other hand, dCas9 may be used to induce gene expression through the activation domains of several transcription factors.
These strategies can be used to validate gene function, direct cell-fate conversion, reprogramming cell to pluripotency and further down the line as a therapeutic approach against various diseases.
Targeted Chromatin Modifications Using Cas9
Researchers have been able to target different histone modifiers to specific genomic loci in ESCs and cell lines using dCas9 fusion proteins. The dCas9 method helps to uncover pressing epigenetic questions involving the precise role of each histone modifying enzyme, the interaction between various chromatin modifiers, and the effect of these histone modifications on local chromatin architecture and gene regulation.
In order to target DNA methylation, the dCas9-based system is fused to the catalytic domain of DNMTs to methylate CpG islands. This method has been shown to induce higher methylation changes in larger genomic regions resulting in greater repression of gene expression than other editing tools, such as TALE and ZFP. dCas9 has also been fused to the catalytic domain of TET1 in order to cause DNA demethylation which may result in detectable changes in mRNA expression.
The only drawback to this method is that the changes in methylation pattern that are produced are not always sufficient to induce changes in gene expression. Occasionally, further recruitment of co-factors or additional changes need to be made in order to determine a cause and effect relationship.
Recently, the CRISPR/dCas9 system has been adapted to visualize native genomic loci in living cells by fusing dCas9 with a fluorescent protein. Compared to other imaging techniques, such as FISH, the CRISPR/dCas9 system does not require fixation, allowing investigators to visualize genomic interactions in living cells. By live imaging the cell, investigators are able to monitor the timing and persistence of chromatin configurations and how they correlate with changes in cell function, morphology, and environmental perturbations.
The biggest advantage for the CRISPR/dCas9 system is that it’s able to specifically amplify the intended epigenetic effect by altering the dCas9 and the gRNA. In order to amplify the signal, the dCas9 protein may be fused to multiple copies of an effector domain or several effectors with similar functions may be used. Or, multiple gRNAs that target a DNA region of interest or gRNA can be engineered to include RNA motifs that are recognized by RNA binding proteins. Both are methods that can be used to amplify the intended modification.
Another approach to amplify an epigenetic effect is by attaching GCN4 peptide epitopes separated by linker sequences to the dCas9 protein. The peptide epitopes are recognized by a single chain antibody, which is in turn linked to an effector protein of choice. When the dCas9-GCN4 array reaches it targeted locus the scFv-effector fusion are recruited and amplify the intended effect. These are all methods in which intended modifications may be amplified in a way that is not as easily possible using other genome editing tools.
Future of the CRISPR/Cas9 System
Engineering and interrogating the epigenome using this technology will significantly contribute to the advancement of fields such as regenerative medicine and stem cell biology. The robustness and ease of CRISPR/dCas9 editing tools opens up the possibilities to manipulate cell-fate transitions, dissect different types of epigenetic mechanisms and study diseases with an epigenetic basis.
Source: Pulecio et al., CRISPR/Cas9-Based Engineering of the Epigenome. Cell Stem Cell, 2017. 21, p431-447.
