On the other hand, arginine methylation of histones H3 and H4 promotes transcriptional activation and is mediated by a family of protein arginine methyltransferases (PRMTs). There are 9 types of PRMTs found in humans but only 7 members are reported to methylate histones. They can mediate mono or dimethylation of arginine residues. Based on the position of the methyl group addition, PRMTs can be classified into type I (CARM1, PRMT1, PRMT2, PRMT3, PRMT6, and PRMT8) and type II (PRMT5 and PRMT7). Type II PRMTs are found to be strongly implicated in diseases like cancer.1 For example, PRMT5 plays a role in the repression of certain tumor suppressor genes such as RB tumor suppressors while PRMT7 overexpression is observed in breast cancer. Detection of activity and inhibition of type II PRMTs as well as other HMTs would be important in elucidating mechanisms of epigenetic regulation of gene activation and silencing, as well as benefiting cancer diagnostics and therapeutics.
Start by isolating your histone proteins from your samples of interest, then select an appropriate ELISA kit to detect histone methylation levels.
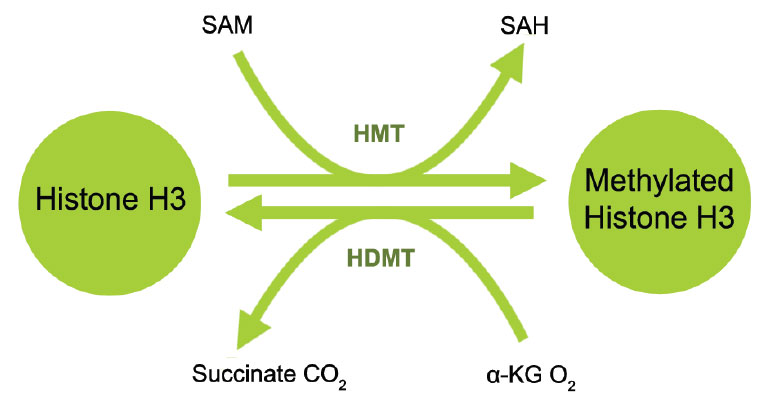
Histone demethylation is the removal of methyl groups in modified histone proteins via histone demethylases. These demethylases have been found to have potential oncogenic functions and involvement in other pathological processes. The discovery of histone demethylases demonstrates that histone methylation is not a permanent modification but rather a more dynamic process. Two major families of demethylases have been discovered: Lysine specific demethylase 1 (LSD1) and Jumonji domain containing (JmjC domain) histone demethylases (JMJD2, JMJD3/UTX and JARIDs). The specific amino acid residue and degree of methylation determines the demethylation enzyme. For example, on histone H3, mono- and di-methylated lysine 4 are demethylated by LSD1 (BHC110, KDM1) and tri-methylated lysine 4 by JARID (1A-1D); di- and tri-methylated lysine 27 are demethylated by JMJD3 and UTX (KDM6A) and mono- and di-methylated lysine 9 are demethylated by JMJD1 and tri-methylated lysine 9 is demethylated by JMJD2.2 Inhibition of histone demethylases may lead to histone re-methylation at specific residues important for chromatin dynamics and gene expression. Furthermore, detection of the activity and inhibition of these enzymes would be important in elucidating mechanisms of epigenetic regulation of gene activation and silencing and may benefit cancer diagnostics and therapeutics.
Extract your nuclear proteins from your sample of interest, then deploy an ELISA-based technique to investigate activity and inhibition levels of histone demethylases of the LSD1 and JmjC-domain families.
Ready to learn about another epigenetic mechanism? Read on: Non-Coding RNA

