Tools of the Trade There are several methods and techniques available for DNA methylation analysis other than using mass spectrometry or HPLC, including: Enzyme Linked Immunosorbent Assay (ELISA). A 5-methylcytosine antibody can also be deployed in an ELISA-based technique to detect global DNA methylation in DNA samples. Wells in a microplate are treated to have high DNA affinity and the methylated fractions of the input DNA are detected using capture and detection antibodies, followed by an absorbance reading with a spectrophotometer.
Bisulfite Conversion DNA bisulfite conversion is a unique tool used to discriminate between unmethylated and methylated cytosine for DNA methylation studies. Only bisulfite modification of DNA followed by PCR amplification, cloning, and sequencing of individual amplimers, yields reliable information on the methylation states of individual cytosines on individual DNA molecules. The bisulfite modification technique uses bisulfite salt to deaminate cytosine residues on single-stranded DNA, converting them to uracil while leaving 5-methylcytosine intact. 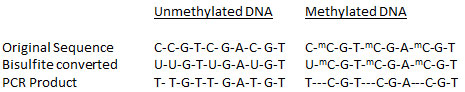 To effectively and efficiently prepare converted DNA for use in various downstream analyses, an ideal DNA bisulfite modification method should be: (1) highly accurate to allow for the complete conversion of cytosine to uracil (correct conversion without deamination of methylcytosine to thymine; and (2) rapid enough to enable the bisulfite process to be as short as possible, since DNA methylation analysis is in high demand for basic research and particularly for clinical applications.
To effectively and efficiently prepare converted DNA for use in various downstream analyses, an ideal DNA bisulfite modification method should be: (1) highly accurate to allow for the complete conversion of cytosine to uracil (correct conversion without deamination of methylcytosine to thymine; and (2) rapid enough to enable the bisulfite process to be as short as possible, since DNA methylation analysis is in high demand for basic research and particularly for clinical applications.
Methylated DNA Immunoprecipitation Highly specific isolation and enrichment of methylated DNA provides an advantage for the convenient and comprehensive identification of methylation status of normal and diseased cells, such as cancer cells. The methylated DNA immunoprecipitation procedure uses an antibody specific to methylcytosine in order to capture methylated genomic DNA. An ideal MeDIP assay should have high sensitivity and specificity, minimal background, and fast high-throughput capability. The enriched and captured DNA can be used for several diagnostic downstream procedures including MeDIP PCR, MeDIP-chip, and MeDIP sequencing and next generation sequencing for genome-wide methylation analysis.
5-hydroxymethylcytosine
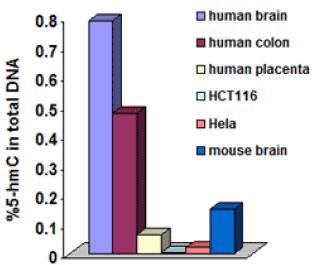
5-hydroxymethylcytosine (5-hmC) is a hydroxylated and methylated form of cytosine. Until the recent discovery of 5-hmC, it was believed that 5-mC was the only DNA base modification. 5-hmC can be generated by the oxidation of 5-mC, a reaction mediated by the TET family of enzymes and DNMT proteins. Early reports suggested the presence of 5-hydroxymethylcytosine in the DNA of bacteriophages and mammalian tissues but these reports could not be confirmed in subsequent studies. 1 It was not until recently (about 2009) that the presence of 5-hmC in mammalian brain cells and mouse embryonic stem (ES) cells was unambiguously proven.2345 These levels were first observed by epigenetics company EpiGentek to vary between different human cell types and tissues and to be significantly decreased in cancer tissues.6
The broader functions of 5-hmC in epigenetics are still unclear today. However, a line of evidence does show that 5-hmC is predominately located within gene promoter regions and is associated with transcriptionally activated genes and plays a role in DNA demethylation, chromatin remodeling, and brain-specific gene regulation. Because of the presence of 5-hmC in DNA with unclear functions in gene regulation and the discovery of the enzymes that produce 5-hmC, it is considered rather important to know the distribution of this base in different cell types and in different compartments of the genome of mammals. Furthermore, it would be particularly important to identify the hydroxymethylation status in human cell/tissues with and without disease if 5-hmC can be proven to have a link between the DNA demethylation process and cancer.
Ready to learn about another epigenetic mechanism? Read on: Chromatin Remodeling

