In arguably the greatest feat of epigenetic orchestration, every single cell in female marsupial and eutherian mammals permanently silences an entire chromosome. While many studied instances of epigenetic regulation involve the repression of a handful of genes, the process of X chromosome inactivation (XCI) effectively silences a 900-gene chromosome over the course of several days and does so for the entire life of the organism (1)(2). The process of entirely yet exclusively silencing one of the largest chromosomes in the human genome continues to baffle researchers, and represents the most ambitious, largest-scale epigenetic operation currently known.
The Origins of X Chromosome Inactivation
Since its discovery in 1961 by Mary Lyon, XCI has been one of the most intriguing epigenetic and evolutionary puzzles in science. The standing hypothesis is that the two sex chromosomes in males and females were autosomes approximately 150-180 million years ago (3). Through the evolution of a sex-determining gene on one of the chromosomes (Sry on what is now the Y chromosome), various inversion events on this chromosome, and subsequent loss of recombination potential, the two chromosomes diverged greatly over the course of a very short evolutionary time period (Figure 1)(4). This massive evolutionary drift between the chromosomes resulted in strong selective pressure for dosage compensation of X-linked genes in female cells. In fact, the equalization of X-linked gene expression between males and females is so effective (and so selected) that dysregulation of the process is lethal.

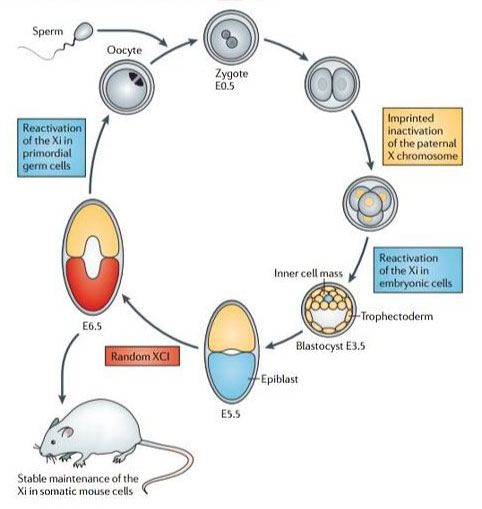
The way in which X chromosome inactivation initiates and is then maintained remains a highly contentious issue. Briefly – eutherian mammals undergo imprinted XCI of the paternal X chromosome at the two to four-cell stage (5). When the embryo is just about to implant (around the 128-cell stage), the epiblast precursor cells in the inner cell mass reactivate, only to randomly inactivate either the maternal or paternal X chromosome shortly thereafter (Figure 2)(4)(5). This dichotomous imprinted XCI in the extra-embryonic tissues and random XCI in the epiblast is then maintained until pregnancy and on through the life of the organism, through countless mitotic cycles (6).
Long non-coding RNAs (RNAs that have been spliced and polyadenylated but not translated) – the genome’s hitmen – play a critical role in both the imprinted and silencing process. The two classically implicated lncRNAs, Xist (Inactive-X Specific Transcript) and Tsix (Xist spelled backwards), are sense and antisense transcripts of the same loci (7). Paradoxically, Xist is exclusively expressed from the inactive X, and covers the inactive X chromosome, shaping it into a condensed ball of chromatin (8). Tsix is only expressed from the active X and represses Xist expression, preventing ectopic silencing of the active X chromosome (9). Imprinted X inactivation initiation is thought to occur by inducing Xist on the paternal X chromosome, while the promoter of the Xist on the maternal X chromosome is methylated, preventing Xist expression (10). This results in exclusive inactivation of the paternal X.
Cats and Future Directions
This marvel of cellular regulation is best seen in calico cats – the gene conferring color is an X-linked gene. Female cats heterozygous for black and orange coat color result in a mosaic pattern of mutually exclusive patches of orange and black (note that the white is due to an autosomal gene that prevents full penetrance of the orange and black colors). This beautifully portrays the early embryonic random X inactivation followed by clonal expansion via countless rounds of mitosis (11).
The elegant mechanism behind this inactivation process remains to be understood. Undaunted, researchers are continuing to interrogate to better understand processes such as aging (12) and mimicking the mechanism to propose cures for Down Syndrome (13).
References
- Wutz A. (2011). Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nature Reviews Genetics, 12:542-553.
- Avner P, Heard E. (2001). X-Chromosome inactivation: counting, choice and initiation. Nature Reviews Genetics, 2:59-67.
- Deakin J, et al. (2009). Unraveling the evolutionary origins of X chromosome inactivation in mammals: insights from marsupials and monotremes. Chromosome Research, 17:671-685.
- Pessia E, Engelstädter J, Marais G. (2014). The evolution of X chromosome inactivation in mammals: the demise of Ohno’s hypothesis? Cell and Molecular Life Sciences, 71:1383–1394.
- Maclary E, et al. (2014). Differentiation-dependent requirement of Tsix long non-coding RNA in imprinted X-chromosome inactivation. Nature Communications, 5:4209, 1-14.
- Chaligné R, Heard E. (2014). X-chromosome inactivation in development and cancer. FEBS letters, 588:2514-2522.
- Gendrel AV, Heard E. (2014). Noncoding RNAs and epigenetic mechanisms during X-chromosome inactivation. Annual Review of Cell and Developmental Biology, 30:561-580.
- Engreitz JM, et al. (2013). The Xist lncRNA exploits three-dimensional genome architecture to spread across the X-chromosome. Science, 341(6147):1237973-1-8.
- Lee JT, et al. (2011). Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nature Reviews: Molecular Cell Biology, 12:815-826.
- Fukuda A, et al. (2014). The role of maternal-specific H3K9me3 modification in establishing imprinted X-chromosome inactivation and embryogenesis in mice. Nature Communications, 5:5464, 1-14.
- Kalantry S. (2011). Recent advances in X-chromosome inactivation. Journal of Cell Physiology, 226(7): 1714-1718.
- Gentilini D, et al. (2012). Age-dependent skewing of X chromosome inactivation appears delayed in centenarians’ offspring. Is there a role for allelic imbalance in healthy aging and longevity? Aging Cell, 11(2): 277-283.
- Jiang J, et al. (2013). Translating dosage compensation to trisomy 21. Nature, 500(7462): 296-300.

